New Genomic Test Shows Promise for Predicting Prostate Cancer Outcomes in Diverse U.S. Veterans
Validation of the Prostatype® P-score for predicting prostate cancer specific mortality in a multiethnic U.S. veterans cohort | Prostate Cancer and Prostatic Diseases
BLUF (Bottom Line Up Front)
A new validation study published in Prostate Cancer and Prostatic Diseases demonstrates that the Prostatype P-score test—which can be performed locally rather than sent to reference laboratories—accurately predicted prostate cancer death in a predominantly African American cohort of U.S. veterans. The test achieved high accuracy (c-index of 0.87) and performed particularly well in intermediate-risk patients, the group that most needs better risk stratification tools. Unlike competing tests, this one can be run in-house at hospitals or certified facilities, potentially reducing wait times and costs while providing critical information to help match treatment intensity to disease aggressiveness.
A Critical Gap in Cancer Care
One of the most challenging aspects of prostate cancer diagnosis is determining which cancers need aggressive treatment and which can be safely monitored. Get it wrong, and patients either receive unnecessary treatment with life-altering side effects, or miss the window to cure a dangerous cancer.
The traditional approach relies on three clinical factors: PSA level, Gleason grade (tumor aggressiveness under the microscope), and clinical stage (how far the cancer appears to have spread). While useful, these factors alone often fail to accurately predict how individual cancers will behave. As the study authors note, "patients with PC often receive mismatched treatments, leading to undertreatment of aggressive disease or overtreatment of indolent disease."
The Science Behind P-score
The Prostatype test examines the expression levels of three specific genes in prostate tumor tissue: IGFBP3, F3, and VGLL3. These are stem cell-related genes that reflect the cancer's biological aggressiveness at the molecular level.
What makes this test unique is its integration strategy. Rather than using gene expression alone, the P-score algorithm combines these three genetic markers with standard clinical information (PSA, Gleason score, and clinical T-stage) to produce a single score ranging from 0 to 15. Higher scores indicate higher risk of death from prostate cancer.
The score divides patients into three risk categories:
- Low risk: 0-2 points
- Intermediate risk: 3-5 points
- High risk: 6-15 points
Dr. Stephen J. Freedland and colleagues at Cedars-Sinai Medical Center and the Durham VA Medical Center set out to determine whether this test, previously validated in Sweden, Spain, and Taiwan, would work in a diverse American population.
Testing in the Real World
The research team identified 1,531 veterans diagnosed with prostate cancer between 2002 and 2019 at the Durham VA. After careful review and tissue quality assessment, 160 patients had sufficient tumor tissue and RNA quality for analysis—a challenge that reflects the reality of working with archived biopsy samples, particularly older ones.
The final study population was 73% Black and 27% White, with a median age of 64.5 years and median PSA of 7.7 ng/mL at diagnosis. This demographic composition is particularly important, as African American men have historically been underrepresented in genomic validation studies despite facing higher prostate cancer incidence and mortality rates.
During a median follow-up of 7.5 years, 51 patients died—14 from prostate cancer and 37 from other causes. This distribution reflects an important reality: many prostate cancer patients die from competing health conditions rather than their cancer, which is why accurate risk stratification matters so much.
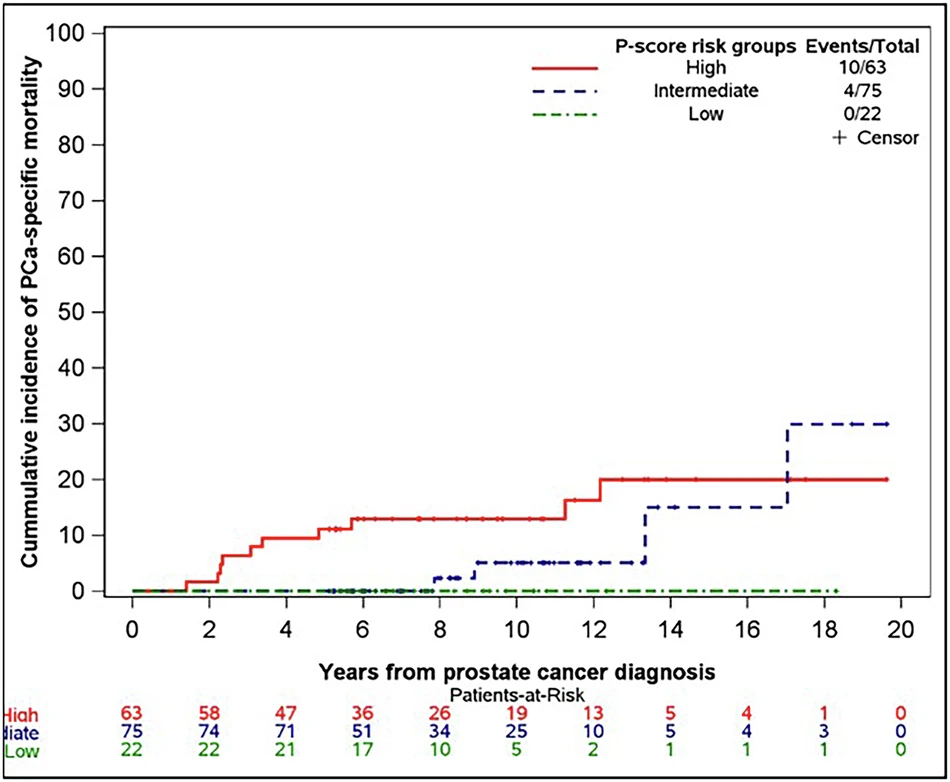
Strong Performance Across the Board
The results were striking. When analyzed as a continuous variable, each one-point increase in P-score was associated with a 48% increase in the risk of prostate cancer death (hazard ratio 1.48, 95% CI: 1.20-1.84, p<0.001).
The test's accuracy metrics were impressive:
- C-index: 0.87 (where 1.0 is perfect prediction)
- 10-year AUC: 0.80 (area under the curve for predicting death within 10 years)
To put this in perspective, these numbers compare favorably with other commercially available genomic tests:
- Decipher: c-index of 0.85
- Prolaris: c-index of 0.78 for 10-year prostate cancer death
- Oncotype DX: time-dependent AUC of 0.84
Critically, P-score outperformed PSA alone, Gleason grade alone, and even the NCCN risk group classification (which already combines PSA, grade, and clinical stage). When the researchers added clinical variables to P-score, the improvement in accuracy was minimal—suggesting the genetic signature provides unique information beyond what standard clinical factors can tell us.
The Intermediate-Risk Challenge
Perhaps the most clinically valuable finding emerged from a focused analysis of intermediate-risk patients. This group faces the greatest uncertainty in treatment decisions: some have indolent disease that could be monitored safely, while others harbor aggressive cancers requiring immediate intervention.
Even when the analysis was restricted to just intermediate-risk patients, P-score remained a highly significant predictor of prostate cancer death (hazard ratio 1.43, p=0.009) with accuracy nearly identical to the full cohort. As the authors note, "intermediate-risk PC presents a great challenge in accurate risk stratification," and these results suggest P-score could fill a critical gap.
Real-World Clinical Implications
The decision curve analysis revealed that using P-score to guide treatment decisions provided net clinical benefit across a wide range of risk thresholds—specifically for patients with a 5-50% risk of prostate cancer death. This range encompasses the most common clinical scenarios where doctors and patients struggle with treatment decisions.
The study identified a sobering reality: six patients (9.5%) with high P-scores were managed with either no treatment or active surveillance. Several of these patients experienced poor outcomes, suggesting their clinical parameters had underestimated how aggressive their cancers truly were. This highlights P-score's potential value in identifying patients at risk of undertreatment.
The In-House Advantage
A key differentiator for the Prostatype test is that it can be performed locally in hospital molecular laboratories (in Europe) or CLIA-certified facilities (in the U.S.), rather than being sent to centralized reference laboratories like competing tests. This design should enable faster turnaround times and potentially lower costs.
A previous Swedish health economics study found that using P-scores was associated with improved quality-adjusted life years while actually lowering overall healthcare costs—though this finding would need confirmation in the U.S. healthcare system with its different cost structure.
The test uses standard RT-qPCR technology on formalin-fixed paraffin-embedded (FFPE) tissue from diagnostic biopsies, the same type of preserved tissue already collected during standard prostate cancer diagnosis. A molecular pathologist selects samples with at least 50% tumor content, RNA is extracted, and the gene expression analysis is performed alongside standard housekeeping genes for quality control.
Study Limitations and Future Directions
The researchers were candid about their study's limitations. The sample size was modest (160 patients) with relatively few prostate cancer deaths (14), which limited their ability to perform more complex statistical modeling. The high drop-out rate from the initial 1,531 eligible patients to the final 160 analyzed reflects real-world challenges in genomic testing.
Tissue availability was particularly problematic for older samples. Pre-2012 practice patterns at the Durham VA involved consolidating multiple biopsy cores into single blocks and using more tissue sections for diagnosis, leaving less material for research. Additionally, obtaining high-quality RNA from 20+ year-old samples proved difficult. The study found that newly acquired samples (particularly from 2018-2019) performed much better, suggesting that future implementation would face fewer technical hurdles with current biopsy protocols.
The study population also received heterogeneous treatments—38% underwent radical prostatectomy, 43% received radiation therapy, and others received various combinations of therapies. While this reflects real-world practice, it prevented the researchers from analyzing whether P-score's predictive value varied by treatment approach.
Building on Previous Validation
This American validation study joins a growing body of evidence supporting P-score's clinical utility:
- The original Swedish development cohort (189 patients, diagnosed 1986-2001) established the three-gene signature's ability to predict overall survival and prostate cancer death
- A 241-patient Swedish study demonstrated that combining the gene signature with clinical parameters significantly improved survival prediction
- The P-score algorithm was locked and validated in a 316-patient Swedish cohort (AUC 0.93)
- A 93-patient Spanish cohort showed an AUC of 0.81
- A 92-patient cohort from Taiwan achieved a c-index of 0.90
Together with the current U.S. veterans study, these validations span diverse healthcare systems and ethnic populations, strengthening confidence in the test's broad applicability.
What This Means for Patients
For men newly diagnosed with localized prostate cancer, particularly those in the intermediate-risk category, P-score represents a potential tool to refine treatment decisions. The test appears most valuable in two scenarios:
Avoiding overtreatment: Low P-scores could provide reassurance that active surveillance is appropriate even when some clinical features suggest higher risk.
Preventing undertreatment: High P-scores could identify biologically aggressive cancers that warrant definitive treatment even when clinical parameters alone might suggest a more conservative approach.
The test's ability to be performed in-house could mean faster results—potentially within days rather than the weeks sometimes required for send-out genomic tests. For patients and doctors navigating the anxiety-filled period after diagnosis, faster access to prognostic information could enable more timely, confident decision-making.
Unanswered Questions
Several important questions remain:
-
Cost-effectiveness in U.S. healthcare: While Swedish data suggest cost savings, U.S. healthcare economics differ substantially. Real-world cost-effectiveness studies are needed.
-
Impact on treatment decisions: This study examined predictive accuracy but didn't track how P-scores actually influenced treatment choices or whether they improved patient outcomes when used to guide therapy selection.
-
Performance by treatment type: Does P-score predict outcomes equally well for patients choosing surgery, radiation, or active surveillance? The current study couldn't answer this due to sample size limitations.
-
Optimal use in clinical algorithms: How should P-score be integrated with other decision tools? Should it replace or supplement other genomic tests?
-
Very high-risk disease: The study excluded patients with very high-risk or metastatic disease at diagnosis, leaving P-score's utility in these populations unknown.
The Broader Context
The validation of P-score in a predominantly African American cohort addresses an important gap in precision oncology. Genomic tests developed and validated primarily in European or Asian populations may not perform equally well across all ethnic groups due to genetic variation, environmental factors, and healthcare access patterns.
By demonstrating strong performance in a 73% Black cohort, this study provides evidence that P-score's biological insights translate across ethnic boundaries—a critical consideration for any test intended for widespread clinical use.
The study also highlights ongoing challenges in translational research. The dramatic attrition from 1,531 eligible patients to 160 with analyzable tissue underscores the importance of current tissue handling protocols that preserve material for future molecular analysis. Modern biopsy practices that place each core in a separate container have greatly improved tissue availability for genomic testing.
Looking Ahead
As prostate cancer care continues to evolve toward precision medicine, tools like P-score represent steps toward matching treatment intensity to individual cancer biology rather than relying solely on population-level risk estimates. The test's in-house capability could democratize access to genomic risk stratification beyond major academic centers.
For the Informed Prostate Cancer Support Group community, this research offers both promise and perspective. Promise, because better risk stratification tools mean better-informed treatment decisions. Perspective, because even the best molecular tests provide probabilities, not certainties, and must be interpreted within each patient's unique clinical context, values, and preferences.
The authors conclude that their findings "support the integration of P-scores into clinical workflows for quick, accurate risk stratification of newly diagnosed PC patients, particularly among those with intermediate-risk disease." Whether this integration happens broadly will depend on forthcoming studies addressing cost-effectiveness, clinical decision impact, and comparative effectiveness against other available genomic tests.
For men facing prostate cancer diagnosis, the expanding toolkit of risk stratification methods—including P-score—represents progress toward one of oncology's most important goals: giving the right treatment, at the right time, to the right patient.
References and Sources
-
Mack A, Tran TD, Berglund E, Andriole GL, Alley C, Sisk AE, et al. Validation of the Prostatype® P-score for predicting prostate cancer specific mortality in a multiethnic U.S. veterans cohort. Prostate Cancer and Prostatic Diseases. 2026 Jan 7. DOI: 10.1038/s41391-025-01070-8. Available from: https://www.nature.com/articles/s41391-025-01070-8
-
Peng Z, Skoog L, Hellborg H, Jonstam G, Wingmo IL, Hjälm-Eriksson M, et al. An expression signature at diagnosis to estimate prostate cancer patients' overall survival. Prostate Cancer and Prostatic Diseases. 2014;17:81-90. DOI: 10.1038/pcan.2013.46
-
Peng Z, Andersson K, Lindholm J, Dethlefsen O, Pramana S, Pawitan Y, et al. Improving the prediction of prostate cancer overall survival by supplementing readily available clinical data with gene expression levels of IGFBP3 and F3 in formalin-fixed paraffin embedded core needle biopsy material. PLoS ONE. 2016;11(1):e0145545. DOI: 10.1371/journal.pone.0145545
-
Soderdahl F, Xu LD, Bring J, Haggman M. A novel risk score (P-score) based on a three-gene signature, for estimating the risk of prostate cancer-specific mortality. Research and Reports in Urology. 2022;14:203-217. DOI: 10.2147/RRU.S361930
-
Pang ST, Lin PH, Berglund E, Xu L, Shao IH, Yu KJ, et al. First validation of the Prostatype® P-score in an Asian cohort: improving risk stratification for prostate cancer. BJUI Compass. 2025;6:e70026. DOI: 10.1002/bco2.70026
-
González-Peramato P, Álvarez-Maestro M, Heredia-Soto V, Mendiola Sabio M, Linares E, Serrano Á, et al. Comparing Prostatype P-score and traditional risk models for predicting prostate cancer outcomes in Spain. Actas Urologicas Españolas (English Edition). 2025;49:501788. DOI: 10.1016/j.acuroe.2025.501788
-
Saemundsson A, Xu LD, Meisgen F, Cao R, Ahlgren G. Validation of the prognostic value of a three-gene signature and clinical parameters-based risk score in prostate cancer patients. The Prostate. 2023;83:1133-1140. DOI: 10.1002/pros.24556
-
Howard LE, Zhang J, Fishbane N, Hoedt AM, Klaassen Z, Spratt DE, et al. Validation of a genomic classifier for prediction of metastasis and prostate cancer-specific mortality in African-American men following radical prostatectomy in an equal access healthcare setting. Prostate Cancer and Prostatic Diseases. 2020;23:419-428. DOI: 10.1038/s41391-019-0195-5
-
Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncology. 2011;12(3):245-255. DOI: 10.1016/S1470-2045(10)70295-3
-
Janes JL, Boyer MJ, Bennett JP, Thomas VM, De Hoedt AM, Edwards VD, et al. The 17-gene genomic prostate score test is prognostic for outcomes after primary external beam radiation therapy in men with clinically localized prostate cancer. International Journal of Radiation Oncology, Biology, Physics. 2023;115(1):120-131. DOI: 10.1016/j.ijrobp.2022.07.1850
-
Fridhammar A, Frisell O, Wahlberg K, Berglund E, Röbeck P, Persson S. Prognostic testing for prostate cancer—a cost-effectiveness analysis comparing a prostatype P-Score Biomarker approach to standard clinical practice. PharmacoEconomics. 2025;43:509-520. DOI: 10.1007/s40273-024-01456-1
-
Cucchiara V, Cooperberg MR, Dall'Era M, Lin DW, Montorsi F, Schalken JA, et al. Genomic markers in prostate cancer decision making. European Urology. 2018;73(4):572-582. DOI: 10.1016/j.eururo.2017.10.036
-
National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: Prostate Cancer. Version 2.2026. Available from: https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf
-
Partin AW, Kattan MW, Subong EN, Walsh PC, Wojno KJ, Oesterling JE, et al. Combination of prostate-specific antigen, clinical stage, and Gleason score to predict pathological stage of localized prostate cancer. A multi-institutional update. JAMA. 1997;277(18):1445-1451. DOI: 10.1001/jama.1997.03540420041027
-
Austin PC, Lee DS, Fine JP. Introduction to the analysis of survival data in the presence of competing risks. Circulation. 2016;133(6):601-609. DOI: 10.1161/CIRCULATIONAHA.115.017719
About the Study Sponsor: This research was sponsored by Prostatype Genomics AB (Nacka Strand, Sweden), the company developing the Prostatype test. Two study authors (Emelie Berglund, PhD and Gerald L. Andriole, MD) are affiliated with Prostatype Genomics. The study underwent independent peer review prior to publication in Prostate Cancer and Prostatic Diseases, a Nature portfolio journal. Open access funding was provided by SCELC (Statewide California Electronic Library Consortium).
For More Information:
- Prostate Cancer and Prostatic Diseases journal: https://www.nature.com/pcan/
- National Comprehensive Cancer Network (NCCN) Prostate Cancer Guidelines: https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf
- American Urological Association: https://www.auanet.org/
- Prostate Cancer Foundation: https://www.pcf.org/
Article prepared for the Informed Prostate Cancer Support Group (IPCSG) Newsletter, January 2026. This article is for educational purposes and should not replace consultation with qualified healthcare professionals.

Comments
Post a Comment